RGS2 protein expression may explain reduced sense of smell in COVID
In a recent study posted to the medRxiv* pre-print server, researchers tested the hypothesis that anosmia in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected individuals elevates the expression of the odorant receptor, a regulator of G protein signaling 2 (RGS2) in humans.
 Study: Elevated expression of RGS2 may underlie reduced olfaction in COVID-19 patients. Image Credit: Nenad Cavoski / Shutterstock
Study: Elevated expression of RGS2 may underlie reduced olfaction in COVID-19 patients. Image Credit: Nenad Cavoski / Shutterstock
Background
A recent study suggested that anosmia during COVID-19 is a consequence of the downregulation of olfactory receptors. Because nasal olfactory neurons do not express proteins needed for SARS-CoV-2 infection, olfactory deficiency during coronavirus disease 2019 (COVID-19) does not directly damage them. Overall, the exact biological mechanisms governing the frequent and sometimes persistent anosmia in COVID-19 are not understood.
About the study
In the present study, researchers collected nasopharyngeal specimens from COVID-19-positive patients and controls not infected with SARS-CoV-2.
First, the team obtained data from the gene expression omnibus (GEO) database of the national center for biotechnology information (NCBI). Next, they analyzed it using ribonucleic acid (RNA)-sequencing to compare the expression of RGS2 in SARS-CoV-2-positive and negative specimens.
The researchers applied a generalized linear model and reported findings for p-value adjusted (Padj)<0.05 in their statistical analysis. In addition, they computed Spearman correlation to identify genes having a strong correlation with RGS2 expression and used Gene Ontology enRIchment anaLysis and visuaLizAtion tool (Gorilla) for gene enrichment analysis.
Study findings
The study analysis showed that SARS-CoV-2-positive patients had upregulated RGS2 expression, which caused anosmia in COVID-19 patients. In addition, the authors observed a 14.5-fold increased RGS2 messenger RNA (mRNA) expression in nasopharyngeal swabs of COVID-19 patients, with Padj=0.001693.
Furthermore, the authors observed that RGS2 expression was associated with the expression of prostaglandin-endoperoxide synthase 2 (PTGS2), interleukin 1 beta (IL1B), C-X-C motif chemokine ligand 8 (CXCL8), and nicotinamide phosphoribosyltransferase (NAMPT) genes. While RGS2 expression was also associated with the expression of other genes encoding COVID-19-induced immune signaling and inflammation markers; however, the correlation was strongest for CXCL8, which encoded interleukin-8 (IL-8).
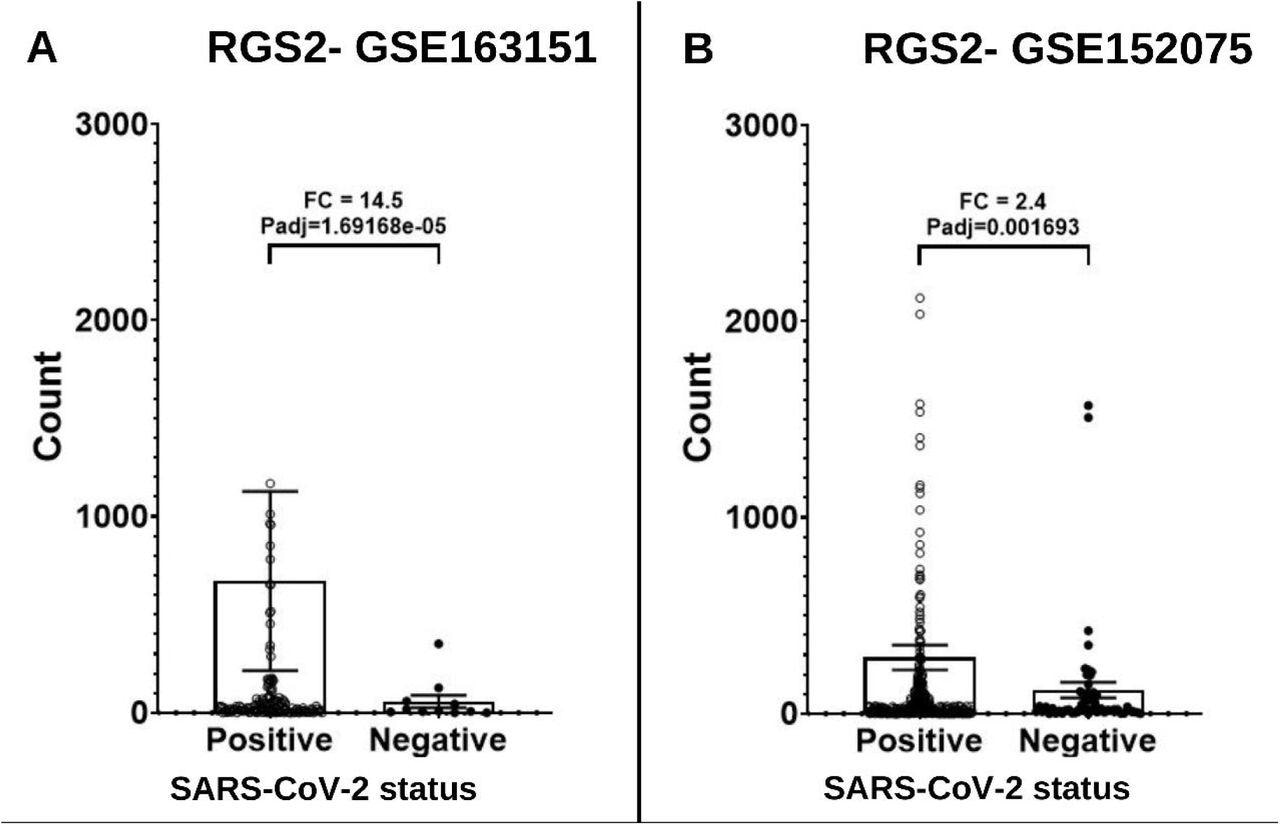
Expression levels of RGS2 mRNA from RNA-sequencing of nasopharyngeal swabs of SARS-CoV-2 positive patients and negative controls. (A) Dataset GSE163151: higher expression of RGS2 (FC= 14.5, padj=1.69e-5) in SARS-CoV-2 positive patients (138 samples) vs. negative controls (11 samples). (B) Dataset GSE152075: higher expression of RGS2 (FC= 2.4, padj=0.0017) in SARS-CoV-2 positive patients (377 samples) vs. negative controls (54 samples). Samples with over 3000 RGS2 counts (five and nine SARS-CoV-2 positive patients, respectively) are not shown due to scale limitations but are included in the statistics.
Studies in small animals have also indicated that the inflammation of olfactory neurons triggered via IL1B signaling caused RGS2 upregulation. The nasal IL1B and RGS2 upregulation during preliminary SARS-CoV-2 infection further confirmed the indirect inflammation hypothesis.
Another study conducted among British Omicron-infected COVID-19 patients showed a 16.7% reduction in anosmia cases. Its findings suggest that the reduced immunogenicity of SARS-CoV-2 Omicron caused a milder immune response, weaker RGS2 expression, and lower risk of anosmia.
The observed changes in gene expression might also put in perspective the corresponding changes in the expression of exosomal microRNAs during COVID-19. Moreover, RGS2 gene expression was associated with accentuated cellular response to IL-8, neutrophil aggregation, and G protein-coupled receptor activity.
Conclusions
Anosmia is one of the earliest and most common symptoms of COVID-19. Of all the demographic groups, young females exhibit the highest RGS2 expression, and therefore, COVID-19-induced anosmia is much more prevalent in young females.
The current study demonstrated that during the first few hours of getting infected with SARS-CoV-2, when anosmia manifests, RGS2 expression is upregulated, thus confirming the correlation between anosmia and RGS2 signaling.
Future research should investigate the cellular mechanisms underlying SARS-CoV-2 infection-driven upregulation of the RGS2 gene and other genes encoding inflammatory markers. These insights could help better understand anosmia and other neurological symptoms of COVID-19.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Elevated expression of RGS2 may underlie reduced olfaction in COVID-19 patients, Eden Avnat, Guy Shapira, David Gurwitz, Noam Shomron, medRxiv pre-print 2022, DOI: https://doi.org/10.1101/2022.05.12.22274991, https://www.medrxiv.org/content/10.1101/2022.05.12.22274991v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Anosmia, Biotechnology, Chemokine, Coronavirus, Coronavirus Disease COVID-19, covid-19, Gene, Gene Expression, Genes, Immune Response, Inflammation, Interleukin, Ligand, Nasopharyngeal, Neurons, Olfaction, Omicron, Protein, Protein Expression, Receptor, Research, Respiratory, Ribonucleic Acid, RNA, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article
