A library of mouse genes to look up the best liver cancer treatment
If you want to get smarter, the library is a good place to start. And for cancer researchers, smarter treatment selection for patients may now start with a library of cancer genes.
In a study published this month in Clinical Cancer Research, a journal of the American Association for Cancer Research, researchers from Osaka University have developed a technique to study a library of genes in lab mice—rather than one specific gene at a time—to identify which cancer genes drive specific liver cancers. There’s hope that this can in turn be used to improve prognostics and guide treatment selection for this deadly cancer.
Hepatocellular carcinoma (HCC), a type of cancer that starts in the liver, is difficult to detect early and to treat, making it one of the top causes of cancer death worldwide. Treatments for HCC do exist, but some work better than others for different people, and it’s hard to know which one to choose. But now, researchers from Japan have developed a method using lab mice that can indicate which liver cancers are driven by specific cancer genes—and what’s more, they’ve found a biomarker that could point out people who will benefit the most from a common HCC treatment.
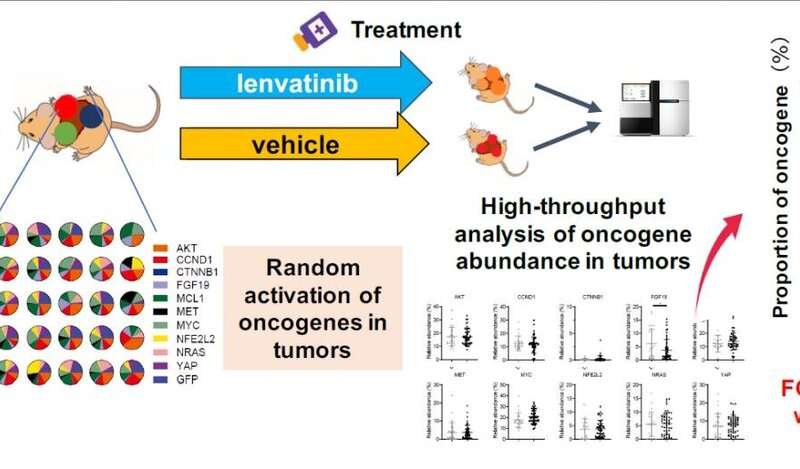
To do this, the researchers first took ten cancer genes that are known to be involved in the pathways leading to HCC and used them to build a DNA “library” by injecting them into lab mice. When the mice developed liver tumors, sequencing the tumor genomes showed which tumors were associated with which genes in the library. Then, mice with liver cancer were treated with the chemotherapy drug lenvatinib to indicate which cancer genes in the library produce tumors that the treatment works particularly well against.
“Our results suggest that FGF19-driven HCC, which generally carries a poor prognosis, may be susceptible to lenvatinib,” says Takahiro Kodama, lead author of the study. “But beyond that specific finding, the technique we used could be used to assess drug susceptibility for other genetic drivers as well.”
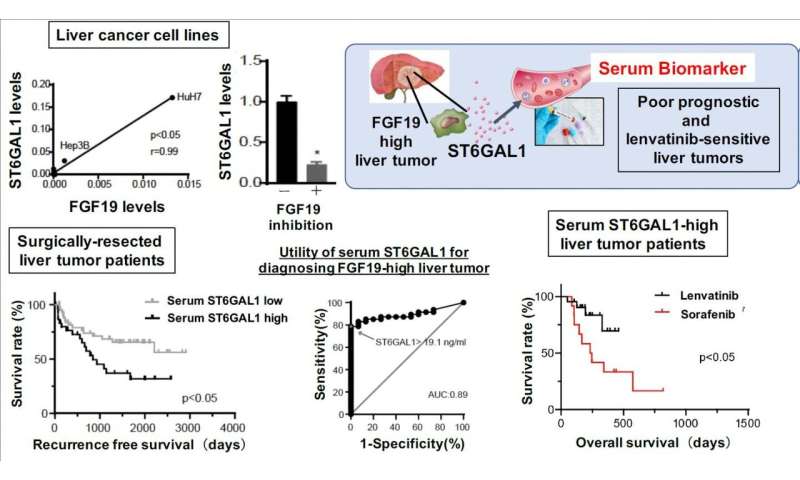
Knowing which gene drives a cancer is powerful information, but to make it useful in the real world of cancer treatment, there needs to be an easy way to identify when patients have FGF19-driven HCC. Working in liver cells in the lab, the research team identified six proteins whose expression was decreased when the FGF19 gene was turned off. From these six, the team determined that a protein known as ST6GAL1 was the most closely correlated with FGF19 in HCC. Further testing of biosamples from actual liver cancer patients confirmed that serum levels of ST6GAL1 could be used to identify patients with FGF19-driven HCC with a high level of sensitivity and specificity.
“While some HCC mouse models already exist, our system can be used to study any gene set, and eliminates the need for expensive and time-consuming genetic studies done one-by-one in individually prepared mice,” says Tetsuo Takehara, senior author of the study. “This new model may be a valuable tool for preclinical drug assessment and increasing the efficacy of drug therapy.”
Source: Read Full Article
