SARS-CoV-2 variants are not evolving to escape from T cell-mediated immunity
In a recent study posted to the bioRxiv* pre-print server, researchers demonstrated that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants did not evolve further to escape from T cell-mediated immunity.
 Study: SARS-CoV-2 variants do not evolve to promote further escape from MHC-I recognition. Image Credit: NIAID
Study: SARS-CoV-2 variants do not evolve to promote further escape from MHC-I recognition. Image Credit: NIAID
Studies have demonstrated that mutations in the SARS-CoV-2 open reading frame 8 (ORF8) gene played a key role in modulating its pathogenesis and adaptation to the host by regulating major histocompatibility complex class I (MHC-I) levels and interferon-stimulated genes (ISGs). Notably, the ORF8 protein induces autophagic degradation of MHC-I and confers resistance to cytotoxic T lymphocyte (CTL) surveillance. Henceforth, during the early phase of the coronavirus disease 2019 (COVID-19) pandemic, studies identified the rapid evolution of the SARS-CoV-2 ORF8 gene.
Several unique mutations within the ORF8 gene have been identified in SARS-CoV-2 variants of concern (VOCs); however, none have increased the ability of ORF8 to suppress MHC-I expression. The MHC-I-induced antigen presentation is crucial for the cluster of differentiation 8 (CD8+) CTL activation, which kills virus-infected cells. Subsequently, researchers speculated that VOC and its ORF8 gene are evolving further to shut down MHC-I evading memory CD8+ T cell-mediated immunity established by prior infection or vaccination.
About the study
In the present study, researchers infected Calu-3 cells with SARS-CoV-2 variants and the ancestral strain (USA-WA1) to examine transcriptional levels of MHC-I genes, which differ for each SARS-CoV-2 variant. They also performed multiple sequence alignment of ORF8 protein sequences from SARS-CoV-2 variants to observe non-synonymous mutations.
Furthermore, the researchers investigated the prevalence of mutations in SARS-CoV-2 variants, for which they downloaded 3,059 SARS-CoV-2 genome sequence data from the global initiative on Avian influenza data (GISAID) database. The team also constructed six ORF8 mutants from SARS-CoV-2 variants to observe the variant-specific effect on the surface MHC-I expression levels of the infected cells.
Lastly, the researchers performed in vivo experiments wherein they infected C57BL/6J mice intranasally with a mouse-adapted (MA) strain of SARS-CoV-2 to analyze the MHC-I expression levels of lung epithelial cells two days after infection.
Study findings
Consistent with previous reports, the USA-WA1 strain significantly downregulated human leukocyte antigen (HLA)-A, B, and C genes. Likewise, the Alpha and Beta VOCs showed a similar reduction in HLA-A, B, and C messenger ribonucleic acid (mRNA) expression as the USA-WA1 strain.
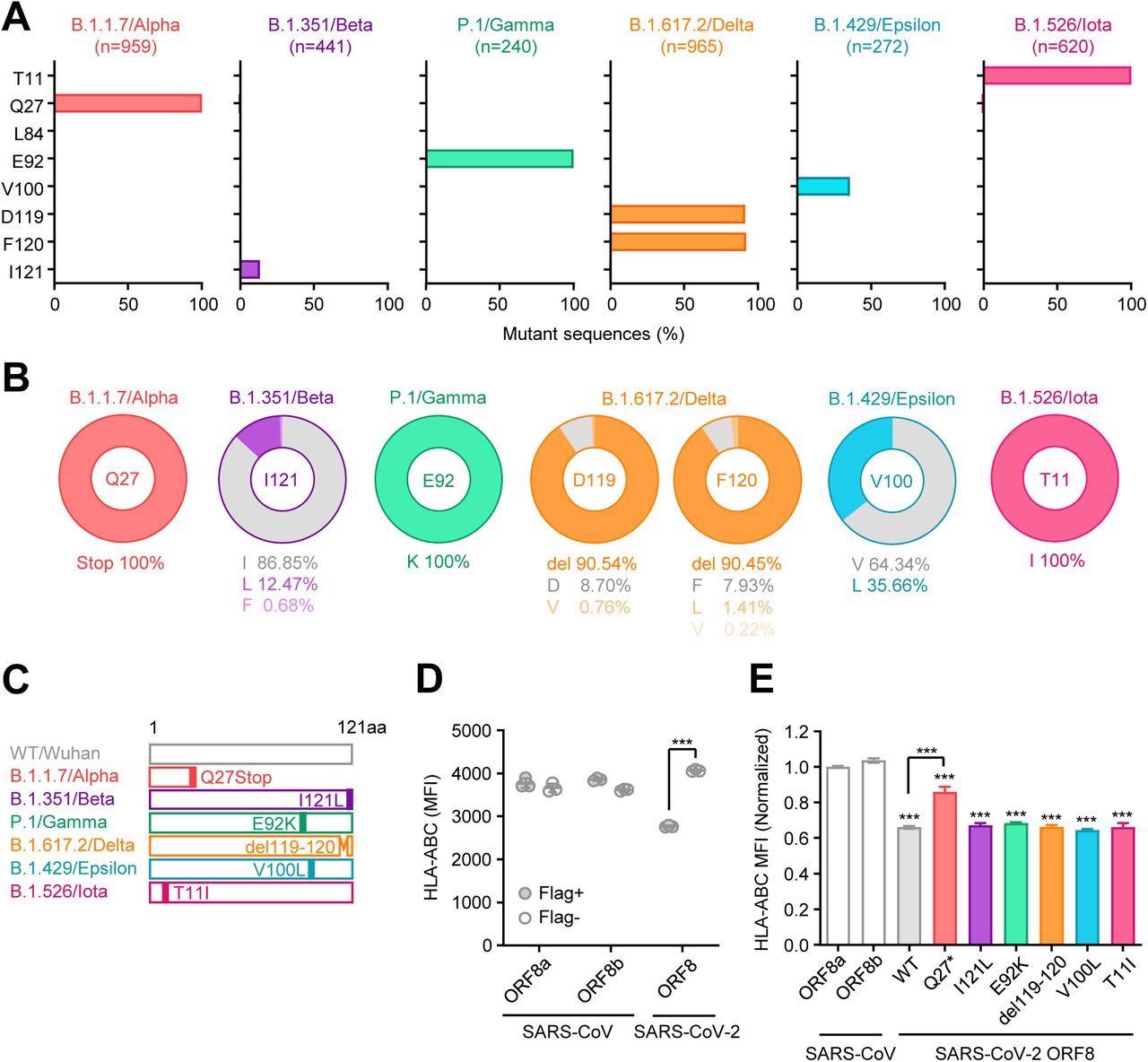
Unique mutations are found in ORF8 gene of SARS-CoV-2 variants (A) Mutant proportion in the ORF8 genes of the indicated SARS-CoV-2 variants. The amino acid positions shown are selected based on the results of multiple sequence alignment performed in Fig.S1. The number of sequences analyzed for each lineage is shown above each graph. (B) Frequency of amino acids at the positions enriched for mutants in each variant. The amino acids shown in gray color correspond to WT. (C) Schematic diagram of ORF8 proteins from SARS- CoV-2 variants. (D and E) HEK293T cells were transfected with plasmids encoding C-terminally Flag-tagged SARS-CoV ORF8a/b, SARS-CoV-2 ORF8 WT, or SARS-CoV-2 ORF8 variants. Forty-eight hours after transfection, cells were collected and analyzed for the cell surface HLA-ABC expression. Data are shown in raw MFI (D) or as the ratio of MFI in Flag+ cells to Flag- cells normalized to the value of SARS-CoV ORF8a (E) (n=3). Data are mean ± s.d. Data are representative of three independent experiments. ***, p< 0.001
Conversely, the Iota and Gamma variants showed weaker downregulation and upregulation of HLA class I genes, respectively. Overall, a majority of SARS-CoV-2 variants reduced HLA transcription just like the ancestral virus. It raised the possibility that many ISGs induced MHC-I processing and presentation pathways were attenuated in Gamma- and B.1.429-infected cells.
Regarding mutations, the authors observed seven non-synonymous mutations and two deletions in the 10 SARS-CoV-2 variants examined. In addition, they identified a premature stop codon at the Q27 amino acid position of the Alpha VOC, which truncated the ORF8 polypeptide length and likely altered its functionality. However, the R52I and Y73C downstream mutations did not impact the Alpha ORF8 protein.
Except for Alpha and Omicron, all other SARS-CoV-2 VOCs, including Beta, Gamma, and Delta, harbored mutations or deletions in the ORF8 protein. Although none of these mutations were conserved among different lineages, SARS-CoV-2 variants of interest, Epsilon, and Iota, had V100L and T11I mutations, respectively.
Furthermore, the authors found several unique lineage-specific mutations. For instance, the ORF8 L84S mutation identified in SARS-CoV-2 clade S was unique and not observed in any other variant. Many mutations discovered via multiple sequence alignment analyses were generally highly prevalent in the proportions ranging from 12.5 to 100%. These findings indicated that the lineage-specific mutations were acquired independently during SARS-CoV-2 evolution, and mutations in a particular amino acid were exclusive to a single lineage. Unlike the influenza A virus, SARS-CoV-2 completely shut down MHC-I induction within infected cells in vivo.
Conclusions
Presumably, SARS-CoV-2, like many other viruses, has developed ways to avoid the efficient MHC-I mediated antigen presentation to CD8+ T cells, as they play a vital role in the host adaptive immune response.
Upon investigating whether SARS-CoV-2 variants shut down the host MHC-I system, the authors discovered that it was not the case rather, the ability to reduce MHC-I expression remained unchanged throughout VOC evolution. Conversely, SARS-CoV-2 VOCs have evolved to limit the host type I interferon response. Therefore, it is highly likely that the SARS-CoV-2 ancestral strain was fully optimized to escape from CD8+ T cell-mediated immunity and was under no evolutionary pressure to further optimize its immune evasion strategy.
This finding also raised the possibility that SARS-CoV-2 utilizes multiple redundant mechanisms to suppress MHC-I expression to ensure escape from CTL killing. Additionally, unlike influenza A or Epstein-Barr virus, it impairs the priming of CD8+ T cells.
Taken together, the study data demonstrated the ability of SARS-CoV-2 to potently avoid the MHC-I-mediated antigen presentation to CD8+ T cells and the role of ORF8 in its efficient replication and transmission.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- SARS-CoV-2 variants do not evolve to promote further escape from MHC-I recognition, Miyu Moriyama, Carolina Lucas, Valter Silva Monteiro, Akiko Iwasaki, bioRxiv 2022, DOI: https://doi.org/10.1101/2022.05.04.490614, https://www.biorxiv.org/content/10.1101/2022.05.04.490614v1
Posted in: Medical Research News | Disease/Infection News
Tags: Amino Acid, Antigen, Avian Influenza, Cell, Codon, Coronavirus, Coronavirus Disease COVID-19, covid-19, Epstein-Barr Virus, Evolution, Frequency, Gene, Genes, Genome, Human Leukocyte Antigen, Immune Response, immunity, in vivo, Influenza, Interferon, Leukocyte, Lymphocyte, Mutation, Omicron, Pandemic, Protein, Respiratory, Ribonucleic Acid, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, T Lymphocyte, Transcription, Transfection, Virus

Written by
Neha Mathur
Neha is a digital marketing professional based in Gurugram, India. She has a Master’s degree from the University of Rajasthan with a specialization in Biotechnology in 2008. She has experience in pre-clinical research as part of her research project in The Department of Toxicology at the prestigious Central Drug Research Institute (CDRI), Lucknow, India. She also holds a certification in C++ programming.
Source: Read Full Article