Novel machine-learning blood test detects cancers with genome-wide mutations in single molecules of cell-free DNA
Novel blood testing technology being developed by researchers at the Johns Hopkins Kimmel Cancer Center that combines genome-wide sequencing of single molecules of DNA shed from tumors and machine learning may allow earlier detection of lung and other cancers.
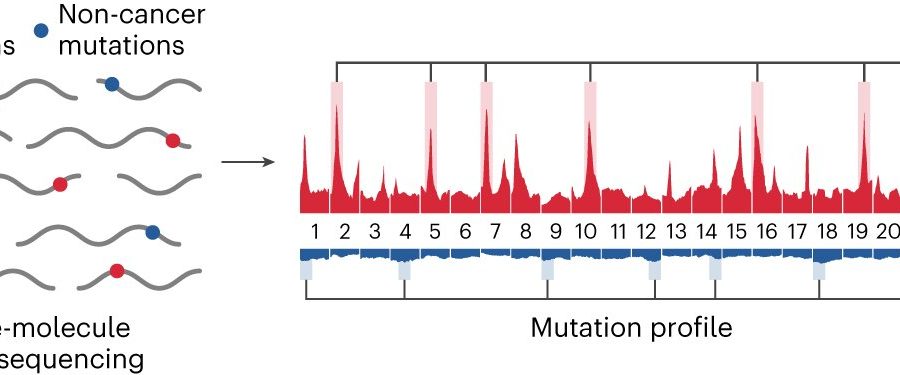
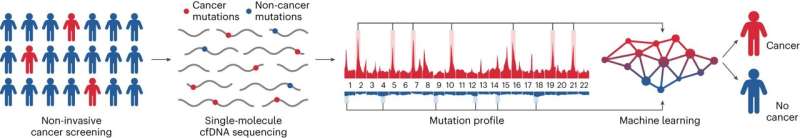
The test, called GEMINI (Genome-wide Mutational Incidence for Non-Invasive detection of cancer), looks for changes to DNA throughout the genome. First, a blood sample is collected from a person at risk for developing cancer. Then, cell-free DNA (cfDNA) shed by tumors is extracted from the plasma and sequenced using cost-efficient whole genome sequencing.
Single molecules of DNA are analyzed for sequence alterations and are used to obtain mutation profiles across the genome. Finally, a machine learning model trained to identify changes in cancer and non-cancer mutation frequencies in different regions of the genome is used to distinguish people who have cancer from those who do not have cancer. The classifier generates a score ranging from 0 to 1, with a higher score reflecting a higher probability of having cancer.
In a series of laboratory tests of GEMINI, investigators found that the approach, when followed by computerized tomography imaging, detected over 90% of lung cancers, including among patients with stage I and II disease. A description of the work, a proof-of-concept study, published online July 27 in the journal Nature Genetics.
“This study shows for the first time that a test like GEMINI, incorporating genome-wide mutation profiles from single molecules of cfDNA, in combination with other cancer detection approaches, may be used for early detection of cancers, as well as for monitoring patients during therapy,” says senior study author Victor Velculescu, M.D., Ph.D., professor of oncology and co-director of the cancer genetics and epigenetics program at the Kimmel Cancer Center.
The study mostly focused on detection of lung cancer in high-risk populations, says Daniel Bruhm, lead study author and graduate student in the human genetics program at the Johns Hopkins University School of Medicine. “However, we detected altered mutational profiles in cfDNA from patients with other cancers, including liver cancer, melanoma or lymphoma, suggesting it may be used more broadly,” Bruhm says.
To develop GEMINI, investigators examined whole-genome sequences of cancers from 2,511 people across 25 different cancers from the Pan-Cancer Analysis of Whole Genomes study, identifying distinct mutation frequencies across the genome in different tumor types. For example, lung cancers were found to have an average of 52,209 somatic mutations per genome.
The investigators also identified genomic regions with the highest number of mutations, finding that genome regions with a high frequency of mutations were similar between tumor tissue and blood-derived cfDNA from patients with lung cancer, melanoma or B cell non-Hodgkin lymphoma.
Researchers evaluated GEMINI’s ability to detect sequence alterations in cfDNA from 365 people in the Longitudinal Urban Cohort Aging Study (LUCAS), a cohort of people at high risk of having lung cancer.
GEMINI scores were higher in people with cancer than in those without cancer. Researchers also assessed whether GEMINI could be combined with DELFI (DNA evaluation of fragments for early interception)—a previously developed test that detects changes in the size and distribution of cfDNA fragments across the genome—to improve detection of early-stage lung cancer.
Several cancer samples that GEMINI missed were detected using the combined technique. In 89 samples from patients from the LUCAS cohort who had lung cancer, GEMINI combined with DELFI correctly detected cancers 91% of the time. Similar results were obtained in a separate validation cohort of 57 people who mostly had early-stage lung cancer.
The investigators also studied the use of GEMINI in other study samples, including seven patients who did not have any detectable tumors at the time of blood collection. They had a median GEMINI score of 0.78, which is higher than people without cancer. Six tested positive using GEMINI and were later diagnosed with lung cancer from 231 to 1,868 days after samples were obtained, suggesting that abnormalities in cfDNA mutation profiles can be detected years before standard diagnoses.
Additional experiments determined that GEMINI could distinguish between subtypes of lung cancers and could detect early liver cancers. In a group of patients receiving lung cancer drug treatment, GEMINI scores decreased during initial response to therapy, suggesting the testing could be used to monitor patients during therapy.
Together, the results indicate that the combination of genome-wide GEMINI mutation analyses and DELFI fragmentation analyses of cfDNA “may provide an opportunity for cost-efficient, scalable detection of cancers,” says Rob Scharpf, Ph.D., associate professor of oncology at the Kimmel Cancer Center. Larger clinical trials are needed to validate the tool before it could become available for clinical use, he says.
More information:
Daniel Bruhm et al, Single-molecule genome-wide mutation profiles of cell-free DNA for non-invasive detection of cancer, Nature Genetics (2023). DOI: 10.1038/s41588-023-01446-3. www.nature.com/articles/s41588-023-01446-3
Journal information:
Nature Genetics
Source: Read Full Article