New framework to identify genetic risk of disease could lead to targeted therapeutics
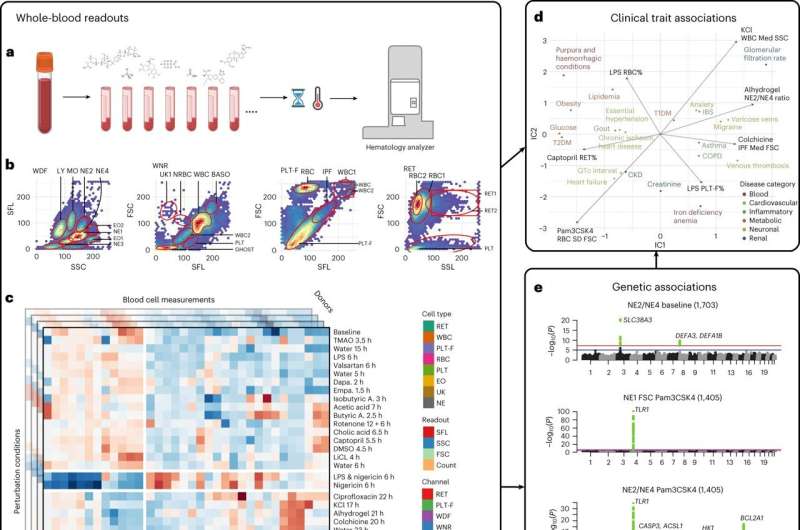
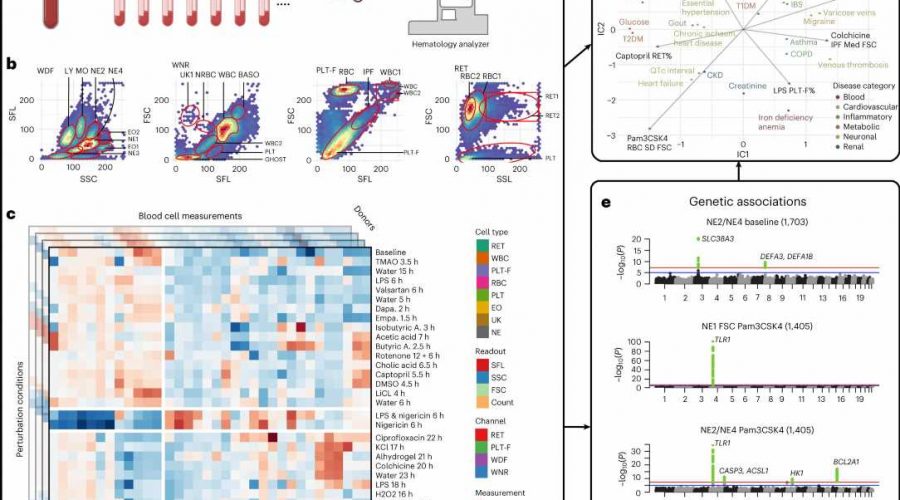
Genome-wide association studies (GWAS) on patient blood samples are useful for identifying the genetic basis of blood cell traits and their links to common diseases. While previous experiments have focused on characterizing clinical parameters such as cell count, few have evaluated the dynamic effects of factors—such as inflammation, microbiome or medications—on blood cell contributions to disease development and progression.
This lack of insight into underlying biological mechanisms behind such chronic progressive conditions has hindered the advancement of targeted therapeutics.
Researchers at Brigham and Women’s Hospital developed a framework to identify potentially hidden genetic contributors to disease by applying different stress tests to human blood cells. Using a range of physical, chemical and pharmacological stimuli, the investigators recorded evoked cellular responses and genetic variants associated with them in nearly 2,600 individuals.
The paper is published in the journal Nature Genetics.
The team found links between a range of blood-response characteristics and subsets of common diseases and were able to define the genetics underlying these distinct subsets of disease. In one example, they utilized the framework to identify and demonstrate a population of activated neutrophils (white blood cells) that can contribute to inflammation in cardiometabolic diseases.
These findings expand the clinical measures currently available to genetically map subtypes of complex diseases.
“We were able to build on existing technology to identify new disease-associated traits,” said author Calum A. MacRae, MD, Ph.D., of the Division of Cardiovascular Medicine. “These tools, when combined with AI, can help improve the classification of common diseases and bring drug discovery into the clinic.”
More information:
Max Homilius et al, Perturbational phenotyping of human blood cells reveals genetically determined latent traits associated with subsets of common diseases, Nature Genetics (2023). DOI: 10.1038/s41588-023-01600-x
Journal information:
Nature Genetics
Source: Read Full Article