Immune cells present long before infection can predict flu symptoms

St. Jude Children’s Research Hospital scientists, in collaboration with the Institute of Environmental Science and Research (ESR) Limited, found that immune cells present in people months before influenza (flu) infection could more accurately predict if an individual would develop symptoms than current methods which primarily rely on antibody levels.
The study found certain immune cells were associated with increased protection, while other immune cells were associated with increased susceptibility to developing symptoms after catching the virus. The findings have implications for new approaches to public health and were published today in Nature Immunology.
“We’ve been struggling for decades, if not centuries, with why some people get sick with infections and some don’t,” said co-corresponding author Richard Webby, Ph.D., St. Jude Department of Host-Microbe Interactions. “This is one of the best attempts to try and figure that out for influenza. We were able to measure many different immune parameters from a single blood draw and correlate them with protection from, or susceptibility to, infection symptoms.”
Functional diversity improves anti-influenza immune performance
The researchers found that having a more functionally diverse set of immune cells was correlated with increased protection from flu symptoms. The group identified these cells by comparing the immune cells present in the blood of patients who had symptoms from flu infection to those who were asymptomatic or uninfected. The blood samples, taken up to six months before that flu season, showed very different sets of immune cells in the two groups.
Those without symptoms not only had a more functionally diverse set of immune cells but those cells were also associated with an influenza-specific long-term response, sometimes called the memory response. Patients with symptoms tended to have a more similar set of inflammatory immune cells, which are more likely to be involved in a nonspecific, functionally narrow and short-term response.
The analysis included volunteers in the surveillance for a community cohort-based influenza-like illness (SHIVERS-II) study in New Zealand. SHIVERS-II includes a unique cohort of volunteer patients that the study tracks over time, including their health information. For this study, the volunteers regularly had their blood drawn so the scientists could characterize their immune cells and find which were associated with protection from flu symptoms.
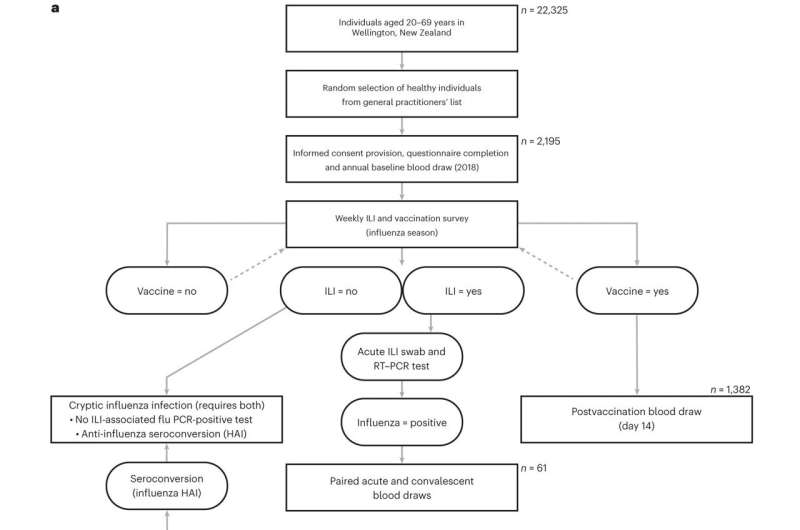
“The SHIVERS platform, which represents a long-running collaboration between St. Jude and ESR, has been tremendously successful because of the willingness of participants to stay engaged in the study,” said co-corresponding author Sue Huang, Ph.D., principal investigator for SHIVERS-II and the World Health Organization National Influenza Centre director at ESR. “It is great to see their efforts coming to fruition.”
“Our results show that the balance of different immune cells in people can be extremely biased,” said senior and co-corresponding author Paul Thomas, Ph.D., St. Jude Department of Immunology. “You might build up an immune cell army that is exceptional at fighting off one kind of infection, but then that can make you feel sicker from another kind of infection. By understanding which immune cells are the best for fighting the flu, we can start designing vaccines to push for those populations that are most protective.”
“The baseline immune state before vaccination is known to significantly vary across age, sex, vaccination status, infection history and more,” said co-first author Aisha Souquette, Ph.D., St. Jude Department of Immunology. “By understanding the different types of immune profiles that can provide protective responses, we can tailor and optimize our vaccine platforms for populations with distinct baseline immune states.”
For developing future tailored approaches, pushing for a particular type of cell or particular immune proteins, such as antibodies, is less important than evaluating the collective contributions of all the immune cells, which may be easier than current methods.
“We observed that the protective, or susceptibility, cell profile’s makeup is less important than the overall, often converging, function,” said co-first author Robert Mettelman, Ph.D., St. Jude Department of Immunology. “This means that we can more broadly evaluate protection or susceptibility at the level of a cell profile, making it easier to evaluate across studies.”
Indeed, this study showed that those vaccinated for the flu generally had increased protective anti-flu immune cells, improving their chance of avoiding symptoms. Those rarer individuals who were unvaccinated and avoided symptoms seemed to have a set of immune cells that mimicked the functions of the protective cells in the vaccinated population.
This may explain why some people are less affected by the flu, even when unvaccinated, than others, but it still suggests vaccination creates the best chance of avoiding symptoms. One way to encourage this vaccine uptake is to determine the inherent risk in staying unvaccinated accurately.
Improving risk predictions by adding to antibodies
With the cell types associated with protection or susceptibility from flu now identified, the future promises improved influenza risk prediction. Clinicians and vaccine developers have traditionally predicted influenza susceptibility by looking at anti-flu antibodies in the bloodstream. Antibodies are proteins that interfere with a virus’s ability to infect cells.
They are produced by a special kind of immune cell called a B-cell. This study shows the presence of a diverse set of immune cells, with a high proportion of helper T cells, a cell involved in the long-term immune response that “helps” B cells, was more predictive of protection than looking at antibodies alone.
“We were surprised that just by identifying the particular cellular populations and combing it with the serological antibody data, we could make such robust predictions,” Thomas said. “We may have the tools to understand susceptibility to infection already in our hands. We can only do it in the lab now, but it’s surprising and potentially exciting that, someday, we may get to a point where we can easily identify at-risk people and provide targeted support.”
In addition to the robustness of predictions from looking at cell types, the timing was also unexpected. The samples were taken up to six months before flu infection occurred but still produced strong predictions, opening new possibilities for public health.
“The exciting thing is that we could identify a subset of people at the beginning of the flu season that may be more likely to get symptomatic influenza,” Webby said. “We could predict who may be at risk well before the virus is even in the population. I don’t think we’ve really been in a position to even think about that before—this could open up new opportunities to prevent flu-based morbidity.”
While influenza researchers may now have new opportunities in flu prevention revealed by the study, its results also confirmed a long-standing message from virology and immunology.
“Our results reemphasize that vaccination prevents influenza symptoms, and now we can point to the increased levels of those immune cells correlated with that protection,” said Thomas. “Get your annual flu vaccine.”
More information:
Robert C. Mettelman et al, Baseline innate and T cell populations are correlates of protection against symptomatic influenza virus infection independent of serology, Nature Immunology (2023). DOI: 10.1038/s41590-023-01590-2. www.nature.com/articles/s41590-023-01590-2
Journal information:
Nature Immunology
Source: Read Full Article
