Combined clinical and genomic data better predicts COVID-19 severity
The current COVID-19 outbreak is one of the worst pandemics in history and has claimed millions of lives across the globe, overwhelmed healthcare systems, and created unprecedented socio-economic ramifications.
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China, in late 2019 and rapidly spread across the world via human-to-human transmission. Infected individuals are either asymptomatic or exhibit a wide range of symptoms ranging from a mild flu-like illness to rapid progression to pneumonia and sometimes death due to respiratory failure. While patients with mild symptoms recover quickly, those with severe COVID-19 require medical intervention, hospitalization, and even ventilation in ICUs.
This heterogeneity in COVID-19 pathogenesis poses challenges for developing treatment protocols. Studies show that severe symptoms of infectious diseases such as COVID-19 are usually associated with host genome variations. Many recent studies, such as genome-wide association studies (GWAS) of severe COVID-19, have revealed the biological mechanisms behind severe COVID-19 requiring hospitalization. In addition to genomic factors, demographics, lifestyles, socio-economic factors, and comorbidities may also affect the exposure, transmission, severity of disease, and viral load. Rigorous analysis of clinical and genomic datasets is essential to find the biomarkers associated with severe COVID-19.
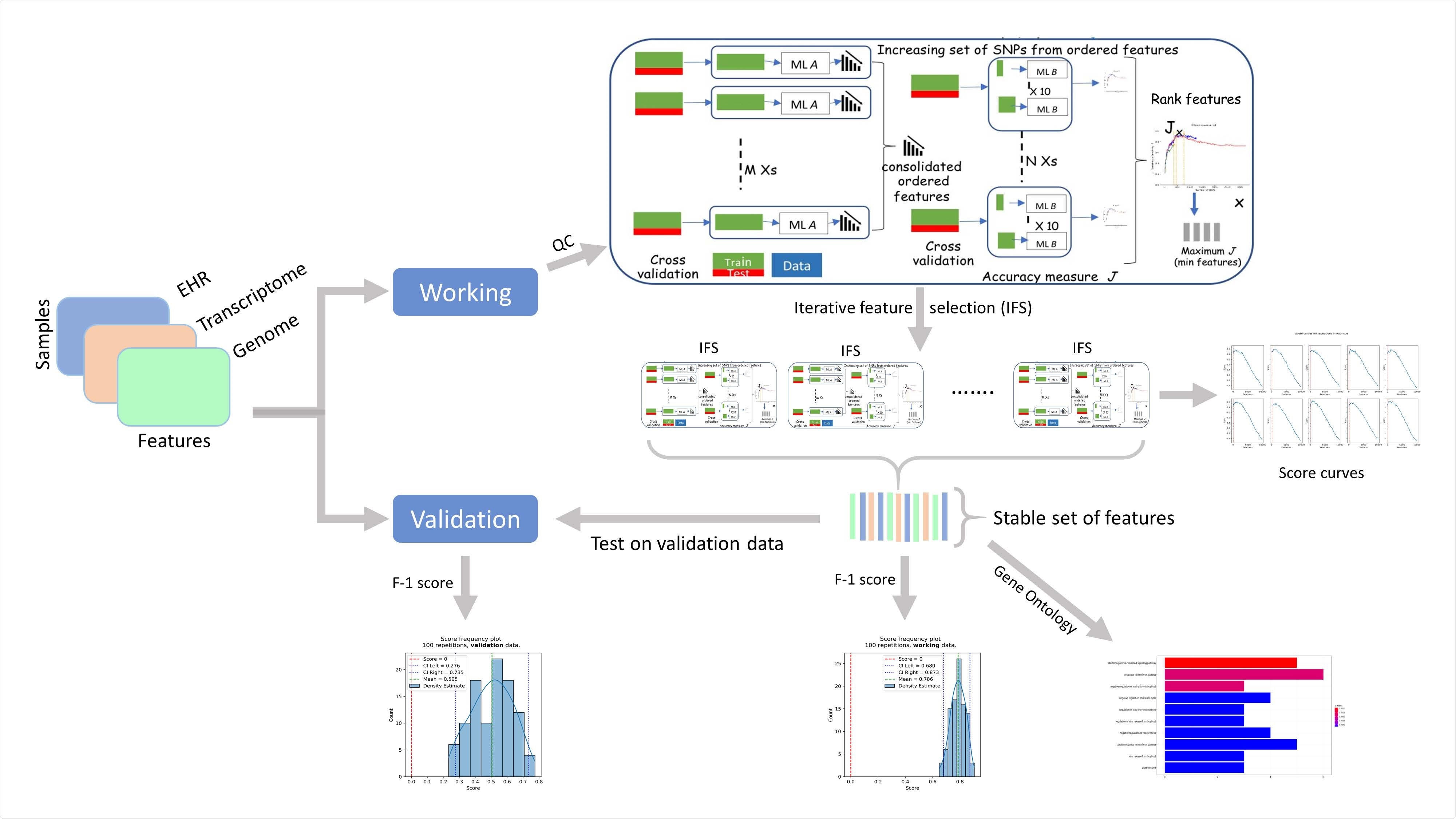
Analyzing clinical as well as genomics data from a large EHR dataset to find the common risk factors in COVID-19 severity
Most studies so far aimed at finding the common risk factors for severe COVID-19 focused either on the clinical or on the genomics factors, but not both of them. To fill this gap, a group of researchers from the US recently analyzed diverse factors from clinical as well as genomics data from a large EHR dataset to find the common risk factors in COVID-19 severity. The study is published in the preprint server, medRxiv*.
The researchers applied AI methods to better interpret the factors that drive the severity of COVID-19. They used the UK BioBank dataset and analyzed both clinical and genomic data of COVID-19 patients, and studied a large prospective cohort of UK patients in the UKBB10 repository. The dataset had a variety of information of about half a million patients related to diagnosis, demographics, lab tests, medications, and genomic information of approximately half a million patients. The data used was from March 16, 2020, to January 21, 2021.
They leveraged positive-unlabeled machine learning algorithms and used a state-of-the-art genomic analysis framework called RubricOE for genomic feature extraction. The results proposed severity prediction algorithms with a high F1 score and provided insights on clinical and genomic factors affecting the severity of the disease.
Although other studies found factors such as smoking, sex (male), age (older than 65), and pre-existing hypertension, cardiovascular disease, diabetes, respiratory diseases, obesity, kidney diseases, immunosuppression, and cancer to be risk factors for severe disease, this study did not find smoking and presence of respiratory diseases to be significant risk factors for the increase in complications due to COVID-19.
Risk factors associated with COVID-19-related mortality is crucial in future research
The study aimed at identifying significant clinical and genomic factors associated with severe response to the SARS-CoV-2 virus and used the data from a large-scale UK EHR dataset. The team used a particular type of machine learning – positive-unlabeled learning – to address the challenge of noisy class label of severity outcome of COVID-19.
According to the authors, the holistic clinico-genomic modeling used in this study can potentially reveal the effect of such factors in a robust way that can be used as biomarkers for better clinical decision making. They plan to investigate further the effect of vaccination and treatment protocols on the severity of COVID-19 by leveraging the inpatient data in the future. Investigating the risk factors associated with COVID-19-related mortality will also be a crucial topic for future research. The authors also reported on how these risk factors have evolved during the last year of the pandemic with respect to significant events such as the emergence of the new variants, including the B.1.1.7 of the SARS-CoV2 virus.
“In the future, we plan to further investigate the effect of treatment protocols and vaccination on COVID-19 severity leveraging the inpatient data.”
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Impact of Clinical and Genomic Factors on SARS-CoV2 Disease Severity Sanjoy Dey, Aritra Bose, Prithwish Chakraborty, Mohamed Ghalwash, Aldo Guzman Saenz, Filippo Utro, Kenney Ng, Jianying Hu, Laxmi Parida, Daby Sow medRxiv 2021.03.15.21253549; doi: https://doi.org/10.1101/2021.03.15.21253549, https://www.medrxiv.org/content/10.1101/2021.03.15.21253549v1
Posted in: Medical Research News | Disease/Infection News
Tags: Cancer, Cardiovascular Disease, Coronavirus, Coronavirus Disease COVID-19, Diabetes, Flu, Genome, Genomic, Genomics, Healthcare, Immunosuppression, Infectious Diseases, Kidney, Machine Learning, Mortality, Obesity, Pandemic, Pneumonia, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Smoking, Syndrome, UK Biobank, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
